What's new
2024-05-23 Localizatome is now open!
2024-05-23 Information on the oxidative stress-dependent localization changes of 8,055 proteins is available.
2024-05-23 Localizatome is now open!
2024-05-23 Information on the oxidative stress-dependent localization changes of 8,055 proteins is available.
Localizatome (Localize-a-tome) is a database that compiles stress-dependent intracellular localization changes of proteins. We focus on the localization changes of intracellular proteins under pathological or inflammatory stress and have established a system called Intracellular Molecule’s Localization Analysis Systems (IMLAS). This system combines a Venus or EYFP fusion human gene library [1-2], a high-throughput microscopy system, and a machine learning program for analyzing the stress-dependent localization changes of human proteins (Figure 1). Using this system, we individually expressed 10,287 fluorescent protein fusion human proteins in cervical cancer-derived HeLa cells and observed the localization patterns of these proteins before and after stress in the same cells. To retrieve the observed images, users can start a search by entering information in the search box on the top page. Users can search for genes using the gene symbol, gene full name, or FLJ clone ID. Currently, information on the oxidative stress-dependent localization changes of 8,055 proteins is available.
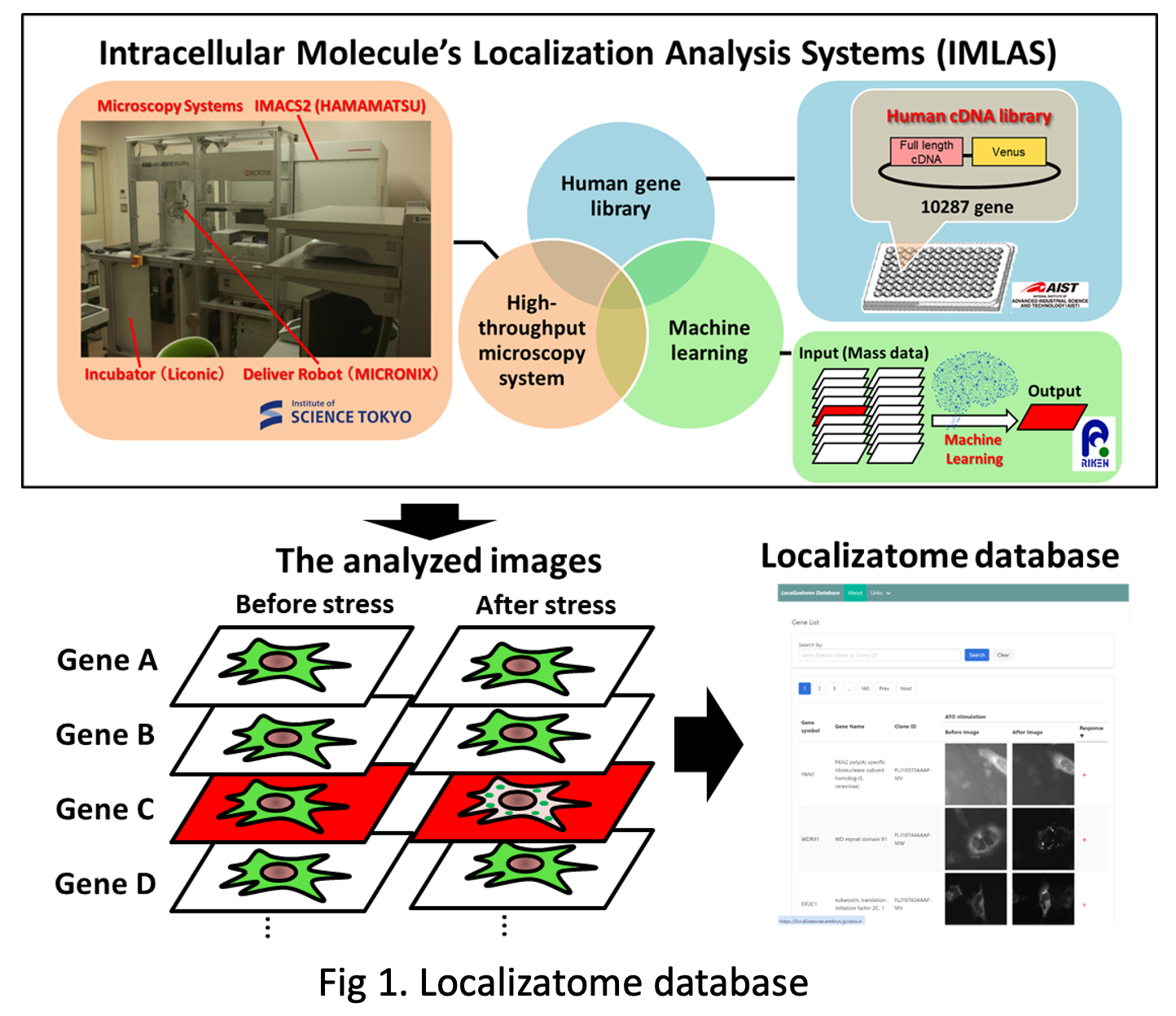
Here, we opted to use a Yellow Fluorescent Protein (YFP) or YFP variant Venus fusion Human Gene Library developed by the National Institute of Advanced Industrial Science and Technology (AIST) [3-5]. In this library Fluorescent Protein is fused at the C-terminal of the human full-length cDNA clones. If the gene had splice variants, we selected the longest variant as the representative gene. We finally chose 10,287 clones from a total of 17,065. For more information, see the Human Gene and Protein Database (HGPD, https://hgpd.lifesciencedb.jp/)
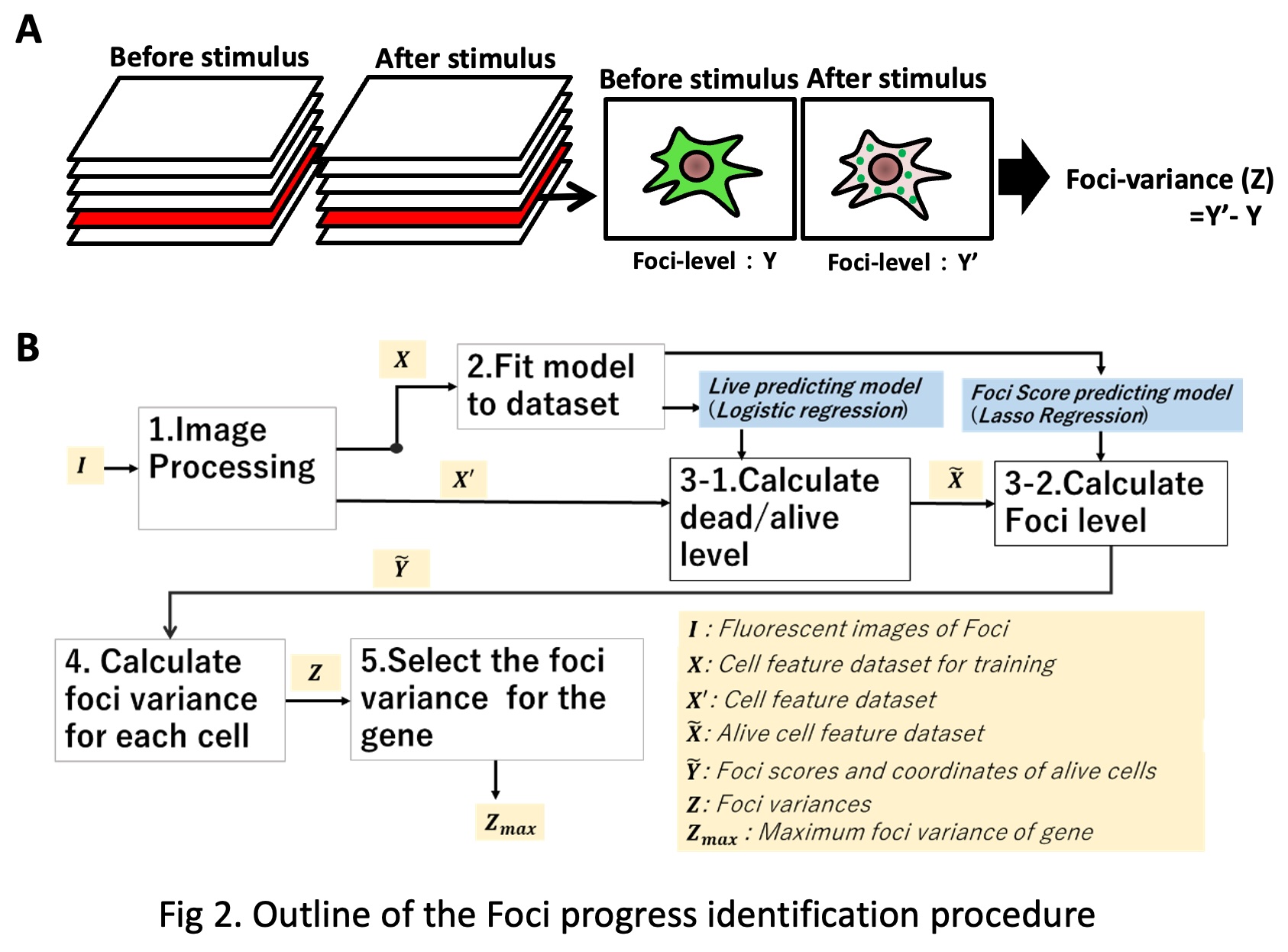
The accumulation of Proteins to intracellular organelles and with other cellular components generates “Foci” – a point of convergence of things, in this case protein particles. These Foci may be the platform of cell signaling and are induced under pathologic or inflammatory stimulus, for example such as in the case of the formation of stress granules. IMLAS, uses Hela cells transfected with the Venus or EYFP fusion human gene library, examines the cells scoring for observed foci formation under oxidative stress induced with arsenic trioxide (ATO) treatment (Fig.2A).
In order to check if whether the foci occur after some gene stimulation, we define some index values: foci-level, foci-variance. When the foci phenomenon progress, we could detect some features like many fluorescent granules in the cell, max intensity and mean intensity are higher. Therefore, we labeled cells in the training dataset foci-level (FL), manually, based on our subjective judgement. FL takes a continuous value within the range of 0≤𝐹𝐿≤4 . FL is a kind of indicator of foci-progress degree. If FL is 0, the cell has no foci features. The bigger FL value is, the more intensive the foci-cell features are.
Foci-variance for cell is the difference of the Foci-levels between before (T001) and after (T002) stimulus. Finally, we identify Foci-variances for each gene by selecting the cell with the maximum Foci variance. Our procedure consists of five parts: 1. Image processing and feature extraction, 2. Model fitting, 3. Scoring Foci-level for Foci-cell, 4. Calculation of Foci-variances for each foci cell before (T001) and after (T002) stimulation, 5. Identification of maximum Foci-variation for gene. The procedure is programmed with Python 3.6 [6]. The outline of the procedure is shown in Figure 2B.
In the Localizatome database, the top five cells with the highest Foci-variance are extracted and outputted. Finally, these five cells were manually assessed to determine whether protein aggregation (Foci formation) changes occurred during oxidative stress, and representative cells with significant changes are publicly available on the web.